de.uni_leipzig.cvk.data.cluster
Class ClusterAlgorithmMCL
java.lang.Object
 de.uni_leipzig.cvk.data.cluster.ClusterAlgorithm
de.uni_leipzig.cvk.data.cluster.ClusterAlgorithm
 de.uni_leipzig.cvk.data.cluster.ClusterAlgorithmMCL
de.uni_leipzig.cvk.data.cluster.ClusterAlgorithmMCL
- All Implemented Interfaces:
- ClusterTableSettings
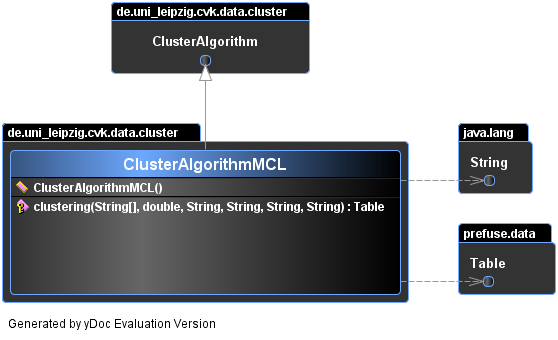
public class ClusterAlgorithmMCL
- extends ClusterAlgorithm
Is a ClusterAlgorithm and an adapt for mcl algorithm of MarkovClustering class.
Implements the clustering(String[], double, String, String, String, String) method and overrides default available settings in abstract class.
-
-
|
Method Summary |
protected prefuse.data.Table |
clustering(java.lang.String[] seeds,
double threshold,
java.lang.String values_A,
java.lang.String values_B,
java.lang.String values_C,
java.lang.String values_D)
An abstract method for a concrete clustering algorithm implementation or adaption. |
| Methods inherited from class java.lang.Object |
clone, equals, finalize, getClass, hashCode, notify, notifyAll, toString, wait, wait, wait |
ClusterAlgorithmMCL
public ClusterAlgorithmMCL()
clustering
protected prefuse.data.Table clustering(java.lang.String[] seeds,
double threshold,
java.lang.String values_A,
java.lang.String values_B,
java.lang.String values_C,
java.lang.String values_D)
- Description copied from class:
ClusterAlgorithm
- An abstract method for a concrete clustering algorithm implementation or adaption.
The clustering method gets the cluster back in a table
with a column name of
ClusterTableSettings.CLUSTER_COLUMN_NAME and with a column type of ClusterTableSettings.CLUSTER_COLUMN_NAME_TYPE.
The column ClusterTableSettings.CLUSTER_COLUMN_SIZE is reserved for a inner sort operation, you don't have to use it, but you can
use this column to add the size of a cluster with type of ClusterTableSettings.CLUSTER_COLUMN_SIZE_TYPE, but be sure that every row is set with a value.
This size defines the order of printing the clusters, the highest value of size for a cluster will print first.
- Specified by:
clustering in class ClusterAlgorithm
- Parameters:
seeds - selected nodes that will be used as seedsthreshold - threshold is between 0 and 1 or -1 for all nodesvalues_A - value of config_A arrayvalues_B - value of config_B arrayvalues_C - value of config_C arrayvalues_D - value of config_D array
- Returns:
- table with one or more columns
 de.uni_leipzig.cvk.data.cluster.ClusterAlgorithm
de.uni_leipzig.cvk.data.cluster.ClusterAlgorithm
 de.uni_leipzig.cvk.data.cluster.ClusterAlgorithmMCL
de.uni_leipzig.cvk.data.cluster.ClusterAlgorithmMCL
de.uni_leipzig.cvk.data.cluster.ClusterAlgorithm
de.uni_leipzig.cvk.data.cluster.ClusterAlgorithmMCL
